
Thanks to some eagle-eyed readers, we’ve been alerted to some corrections for high profile stem cell scientist Jacob Hanna that we had missed, bringing our count to one retraction and 13 errata on 10 papers.
The problems in the work range from duplications of images, to inadvertent deletions in figures, to failures by his co-authors to disclose funding sources or conflicts of interest. Hanna is the first or last author on 4 of the papers, and one of several on the rest.
First up, a correction to a Cell paper on which Hanna is the first author:
1. “Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency” has 553 citations, according to Thomson Scientific’s Web of Knowledge.
Here is the correction:
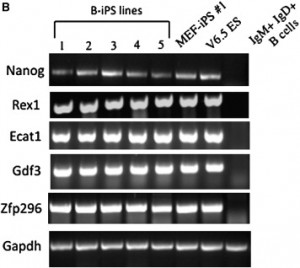
In Figure 4B of the above article, the first lane for measuring Zfp296 gene expression was inadvertently deleted when preparing the final figure; this caused a misalignment of this panel with the other panels in the figure. The modified figure below shows the corrected version for Figure 4B. We apologize for any inconvenience caused by this error.
2. “Novel insights on human NK cells’ immunological modalities revealed by gene expression profiling” was published in The Journal of Immunology. Hanna is the first author of the paper, which has been cited 87 times, according to Thomson Scientific’s Web of Knowledge.
The correction is paywalled — tsk tsk — but here it is in full:
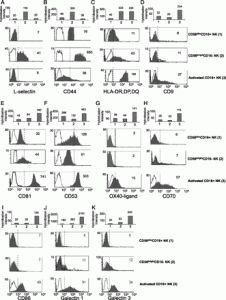
During the preparation of Fig. 3G, a flow cytometry analysis (FACS) plot image was inadvertently duplicated from Fig. 3D (middle panel). We now enclose a corrected Fig. 3 with the validated Fig. 3G. This change does not affect the conclusions or interpretations of findings presented in our article in any way. The figure legend was correct as published and is shown below for reference.
Confirmation of expression patterns of selected differentially expressed genes at the protein level. CD56dimCD16+, CD56brightCD16–, and activated CD16+ NK were sorted and purified as described in Materials and Methods. Each data set (A–K) shows flow cytometry staining for different molecules (light gray) compared with background staining (empty histograms). Dashed lines were added to assist in demonstrating the observed differences in protein expression. Mean fluorescence intensities (MFI) for each sample are indicated in the right upper corner of the panels. MFIs for background stainings were between 2 and 4. Bar graph above each histogram data set represents the normalized hybridization intensity of a probe set specific for the same gene. Data shown are representative stainings obtained from three to five different donors.
3. A Nature article, “Deterministic direct reprogramming of somatic cells to pluripotency,” on which Hanna is the last author, has 144 citations.
Here’s the correction:
In this Article, the reduced representation bisulphite sequencing (RRBS) data obtained and analysed were not initially uploaded to the Gene Expression Omnibus (GEO) record, but can now be found under accession number GSE64115, within the GSE49767 super series. Furthermore, gene expression data for two samples (Mbd3+/+ embryonic stem cells and Mbd3+/+ mouse embryonic fibroblasts (MEFs) after 8 days on doxycycline (dox)) were generated previously by our group1 and deposited in the GEO under accession numbers GSM874650 and GSM874654. Further analysis of this data set was included in Figs 3a, 5c and Extended Data Fig. 8a, c of this Article. Also, in Fig. 5c and Extended Data Fig 8c of this Article, the ‘Mbd3+/+ MEF + 8 day dox’ sample was inadvertently mislabelled as ‘Mbd3+/+ MEF + 11 day dox’. This change does not influence the conclusions of the manuscript.
In addition, further details on the experimental settings used to generate the data in the GSE49766 series reported in this Article (under the GSE49767 super series) have been added to the GEO website. It now states that in Fig. 3a and Extended Data Figs 5 and 8, out of 20 independent clonal series generated in our study carrying either the GOF18 ΔPE-Oct4-GFP transgenic reporter (Addgene plasmid 52382) or the complete GOF18 Oct4-GFP transgenic reporter (Addgene plasmid 60527), the clonal series selected for genomic analysis included anMbd3+/+ clone that carries the GOF18 ΔPE-Oct4-GFP transgenic reporter, and Mbd3flox/− andMbd3−/− cells that carry the GOF18 Oct4-GFP transgenic reporter (complete Oct4 enhancer region with distal and proximal enhancer elements). These reporters can be identified when analysing the Oct4 locus in genomic DNA input datasets. As we do not use Oct4–GFP or any other selection for sorting cells before conducting genomic experiments, the difference in transgene reporters would not influence the interpretation of our genomic analysis data in any way. In these genomics studies, the endogenous Nanog and Oct4 loci are not manipulated and are identical between all cell lines because the Oct4–GFP reporters were introduced via random transgenesis and validated for specificity. Notably, for the mouse induced pluripotent stem (iPS) cell efficiency results presented in our Article (for example, in Figs 1 and 2 and Supplementary Videos 1–4), cell lines carried the matched Nanog–GFP knock-in reporter or Oct4–GFP transgenic reporter (containing both distal and proximal enhancer elements as delineated in Extended Data Fig. 3a). Thus, all iPS cell efficiency and kinetic comparisons were conducted by using matched and validated pluripotency reporter systems.
4. Another Nature article, “Derivation of novel human ground state naive pluripotent stem cells,” on which Hanna is also the last of many authors, has been cited 134 times.
Here’s the correction:
The reduced representation bisulphite sequencing (RRBS) data generated and analysed in this Letter were not originally uploaded to the Gene Expression Omnibus (GEO), but can now be found under accession number GSE52617 (within the GSE52824 super series). As can be seen in this RRBS data, mouse naive pluripotent cells expanded for an extended period (that is, 90 days) in naive 2i/LIF (inhibition of ERK1/2 and GSK3β signalling plus leukaemia inhibitory factor)) conditions, globally lose DNA imprinting.
5. “The mechanisms controlling NK cell autoreactivity in TAP2-deficient patients” was published in Blood and has been cited 44 times. Hanna is one of 19 authors.
Here’s the correction:
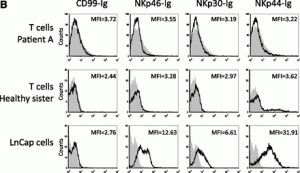
On page 1777 in the 1 March 2004 issue, there are errors in Figure 4. During the preparation of Figure 4B, a flow cytometry analysis plot image was inadvertently duplicated. The negative staining with NKp30-Ig of patient A is duplicated in the NKp44-Ig staining of the healthy sister, and the negative staining of NKp30-Ig of the healthy sister is duplicated in the negative staining of the NKp44-Ig of patient A. This change does not affect the conclusions or interpretations of findings presented in the manuscript. The corrected Figure 4B is shown. The authors apologize for these mistakes.
6. “Harnessing soluble NK cell killer receptors for the generation of novel cancer immune therapy” was published in PLOS ONE. Hanna is one of 12 authors.
The paper has two corrections. Here’s the first:
The funding information for this paper is missing. It should read:This work was supported by the Israel Science Foundation (O.M.), the Binational Science Foundation (O.M.), the AICR (O.M), the ICRF (O.M.), the European Commission (LSHC-CT-2002-518178 and MRTN-CT-2005 to O.M.), the Israel Ministry of Health (A.P.), the Cooperation Program in Cancer Research of the Deutsches Krebsforschungszentrum (DKFZ), and Israeli’s Ministry of Science and Technology (MOST) (A.P.).
And the second:
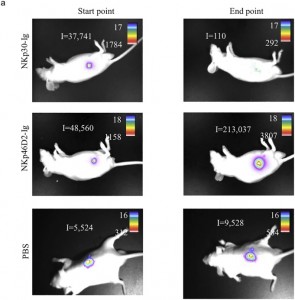
There are errors in Fig 4, “Summary of fusion protein treatment”. Fig 4a contains the wrong set of images for PBS (control) injected mice. Please see the corrected Fig 4 here.
7. Reprogramming of murine and human somatic cells using a single polycistronic vector in PNAS, on which Hanna is on of seven authors, has 265 citations.
There are two corrections.
Here’s the first, which concerns a few typos:
The authors note that on page 157, in the right column, the heading, “Generation of Mouse Ips Cells Using a Single Polycistronic Virus,” should instead read: “Generation of Mouse iPS Cells Using a Single Polycistronic Virus.” On page 158, right column, in line 5 of the first paragraph and line 6 of the second paragraph, ?8should instead appear as ≈8. On page 159, left column, line 9 of the second paragraph, the sentence “This is two orders of magnitude lower than that of ‘secondary’ fibroblasts or B cells carrying preselected DOX-inducible proviruses (19),” should read: “This is one to two orders of magnitude lower than that of ‘primary’ infected fibroblasts (3, 7).” Also on page 159, in the right column, the heading, “Generation of Human IpsCells Using a Single Polycistronic Virus,” should instead read: “Generation of Human iPS Cells Using a Single Polycistronic Virus.” Finally, on page 160, in the first paragraph of Materials and Methods, the sequence for the P2A peptide “GCCACGAAGCAAGCAGGAGATGTTGAAGAAAACCCCGG GCCT” should instead read “GCCACGAACTTCTCTCTGTTAAAGCAAGCAGGAGATGTTGAAGAAAACCCCGGGCCT.”
Here’s the second, regarding an omission of a conflict of interest involving last author Rudolf Jaenisch:
The authors inadvertently neglected to state that, at the time of publication, RJ was an advisor to Fate Therapeutics. We regret this error.
8. A Cell Stem Cell paper, which we’ve previously reported on for having a correction, and then a correction to that correction, contains now a third note, about the same conflict of interest. It refers to another paper that Hanna did not co-author.
Here’s the third note for “Metastable Pluripotent States in NOD-Mouse-Derived ESCs,” which has been cited 184 times and lists Hanna as a first author:
An acknowledgement of potential conflict of interest was inadvertently omitted from this paper. We failed to acknowledge that, at the time of publication, R.J. was a cofounder of Fate Therapeutics. In addition, R.J. served as an advisor to Fate Therapeutics at the time of publication of Hockemeyer et al. (2008).
Update 9/3/15 1:43 p.m. eastern: This post has been updated to clarify that the conflicts of interest stemmed from Hanna’s co-authors, and the corrections were not new, we had simply missed them previously. We also corrected the number of times #2 had been cited, as our original search by DOI gave the wrong result.
Update 9/8/15 12:32 p.m. eastern: A reader (Hanna himself, actually) alerted us to another older correction we had missed, bringing our count to 14 errata.
Like Retraction Watch? Consider making a tax-deductible contribution to support our growth. You can also follow us on Twitter, like us on Facebook, add us to your RSS reader, and sign up on our homepage for an email every time there’s a new post. Click here to review our Comments Policy.
Question – do funding agencies, that evaluate grant applications, ever look at an applicant’s retraction record, as an indicator of someone with a rather flexible interpretation of truth and lies, integrity and fraud?
Slight detail: Article 2 is indexed in Web of Science and has 86 cites. The correction is also there.
“Novel insights on human NK cells’ immunological modalities revealed by gene expression profiling
Fixed that, thanks for flagging. A DOI search didn’t turn up anything, for some reason, but you’re right, searching by title showed 87 cites.
“The problems in his work …., to failures to disclose funding sources or conflicts of interest”.
By reading the corrections listed above, it is clear that Jacob Hanna never failed to do that but it was his previous supervisors. These corrections clearly state “RJ” (Rudolf Jaenisch) or “OM” (Ofer Mandelboim) did not disclose funding or conflict of interest.
We’ve updated the post. Thanks!
Please be accurate and more professional:
– Correction for 2008 cell paper was posted 3 months after publication in 2008 (7 years ago), and is not new.
– 2 Correction for a 2009 PNAS paper are both from 2009 and do not involve Hanna at all (and are not new).
– The Cell Stem Cell note mentioned involves RJ, and is from 2009, and is not new.
“contains now a third note???”
……………
We’ve updated the post. Thanks!
It should be also noted that the 11 year old retracted paper by referred to above by RW, has been corrected and republished on bioRxiv:
http://biorxiv.org/content/early/2015/03/30/016816
8 more publication for Stem Cell Researcher Hanna (in 2015):
1) Geula et al. Science 2015:
http://www.ncbi.nlm.nih.gov/pubmed/25569111
2) Irie et al. Cell 2015:
http://www.ncbi.nlm.nih.gov/pubmed/25543152
3) Manor et al. Curr Opin Genet Dev. 2015:
http://www.ncbi.nlm.nih.gov/pubmed/26291026
4) Maza et al. Nature Biotechnology 2015:
http://www.ncbi.nlm.nih.gov/pubmed/26098448
5) Weinberger et al. Nature Reviews Molecular Cell Biology 2015:
http://www.nature.com/nrm/posters/pluripotency/index.html
6) Shakiba et al. Nature Communications 2015:
http://www.ncbi.nlm.nih.gov/pubmed/26076835
7) Nicenboim et al. Nature 2015:
http://www.ncbi.nlm.nih.gov/pubmed/25992545
8) Zviran et al. BioRxiv 2015:
http://dx.doi.org/10.1101/013961
In addition to 4 corrections involving Lack of disclosure of conflict of interest by Rudolf Jaenisch highlighted above (and NOT Hanna), there are 8 corrections since 2011 by Jaenisch on which Hanna is NOT even an author.
1) http://www.nature.com/ncomms/2015/150730/ncomms9008/full/ncomms9008.html
2) http://www.nature.com/ni/journal/v16/n8/full/ni0815-889a.html
3) http://www.sciencedirect.com/science/article/pii/S1934590914003944
4) http://www.sciencedirect.com/science/article/pii/S1934590914003403
5) http://www.sciencedirect.com/science/article/pii/S2213671114002008
6) http://www.sciencedirect.com/science/article/pii/S0092867413015341
7) http://www.sciencedirect.com/science/article/pii/S1097276513002153
8) http://www.sciencedirect.com/science/article/pii/S009286741100849X
I guess Rudolf Jaenisch beats Hanna according to your count (and only started counting since 2011 for RJ).
Based on your criteria for counting corrections, I believe you have actually missed one correction by my co-authors back from 2008. Always glad to correct 🙂
http://www.nature.com/nature/journal/v454/n7205/full/nature07196.html
” In both the Methods Summary and the online-only Methods (cell culture section), the concentration of AZA is incorrectly listed as 0.5 mM. The correct final concentration is 0.5 microM”.