 A highly cited cancer researcher at MD Anderson has notched three major corrections, all associated with problems in figures. One note cites “human error” as the cause.
A highly cited cancer researcher at MD Anderson has notched three major corrections, all associated with problems in figures. One note cites “human error” as the cause.
Bharat Aggarwal is the last author on all three papers. He is now up to six corrections, two unexplained withdrawals, and two Expressions of Concern. He’s also threatened to sue us in the past, and has told us that his institution has been looking into his work.
Only one note specifies that the correction does not affect the paper’s conclusions.
First up: “Inhibition of growth and survival of human head and neck squamous cell carcinoma cells by curcumin via modulation of nuclear factor-κB signaling,” published in the International Journal of Cancer and cited 168 times, according to Thomson Scientific’s Web of Knowledge. The issues span two figures, according to the erratum note:
Three different concerns were brought to our attention. First, in Fig. 2a control lane at 2 h was reused for control at 4h. Second, loading controls were reused in Fig. 4A, 4B, 4D and 4E (lower beta acitn for cyclin D1, Bcl-2, COX-2 and MMP-9). Third, loading controls were reused in Fig. 4A, 4B and 4E (upper beta actin for cyclin D1, Bcl-2 and COX-2). Following additional information (indicated in italics & bold) to the legends of Figure 2a and Figure 4, provides the explanation for this.
Figure 2 -(a) Curcumin inhibits constitutive nuclear NF-kB in HNSCC. MDA 686LN cells (2 ×106 cells/ml) were treated with the indicated concentration of curcumin for either 2 or 4 hr and then tested for NF-kB by EMSA as described in Material and Methods. A typical of 3 independent experiments is presented. Lane labeled as 0 uM curcumin for 2 hours and 0 uM curcumin for 4 hours were derived from the same treatment.
Figure 4- Curcumin downregulates NF-kB regulated gene products. Whole-cell lysates from 2 x 106 cells/ml (JMAR C42, Tu 167, Tu 686, MDA1986, MDA 686LN) were tested for constitutive expression of Cyclin D1 (a), Bcl-2 (b), Bcl-xL (c), MMP-9 (d), COX-2 (e) by Western blot analysis as described in Material and Methods. To examine the effect of curcumin 5 × 106 cells/ml MDA 686LN cells were treated with indicated doses of curcumin for 4 hr, and the cytoplasmic extracts prepared and resolved on 10% SDS-PAGE gel, electro transferred on to a nitrocellulose membrane, and probed for the following: cyclin D1 (a), Bcl-2 (b), Bcl-xL (c), MMP-9 (d) and COX-2 (e). For Bcl-xL, U266 cells, a human multiple myeloma cell line, was included as a positive control. In each case, the same blot was stripped and reprobed with anti-b-actin antibody to show equal protein loading (lower panel in each figure). Beta actin controls (for different cell lines) in panel (a), (b) and (e) were derived from the same experiment. Beta actin controls (for different curcumin doses) for panel (a), (b), (d) and (e) were also derived from the same experiment. A typical of 3 independent experiments is presented.
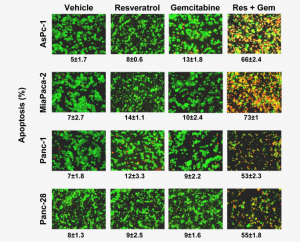
Here’s more from the the corrigendum for “Resveratrol, a multitargeted agent, can enhance antitumor activity of gemcitabine in vitro and in orthotopic mouse model of human pancreatic cancer,” which has been cited 66 times, and was also published in the International Journal of Cancer:
Because of human error discovered in one of the panels in Fig. 1C, experiment was repeated and results are shown below. The results remain unchanged. This correction does not influence the conclusion of any portion of the entire study. The authors regret this error.
Figure 1. Resveratrol inhibits the growth and proliferation, enhances the apoptotic effects of gemcitabine. (c) Live/Dead assay results indicated that resveratrol (Res, 10 uM) potentiates gemcitabine (Gem, 100 nM)-induced cytotoxicity. Data indicated as percentage proportions of apoptotic pancreatic cancer cells. Data are a representative of two independent experiments. Values are mean +/- SD of two independent experiments. The photographs were taken at the magnification of x20.
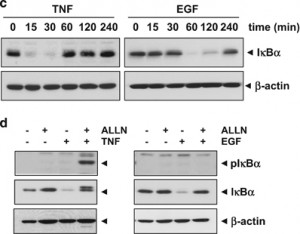
Last but not least, “Epidermal growth factor (EGF) activates nuclear factor-κB through IκBα kinase-independent but EGF receptor-kinase dependent tyrosine 42 phosphorylation of IκBα” was published in Oncogene. The paper has been cited 62 times. Here’s the corrigendum:
Since the publication of the above article the authors have identified that β-actin, the loading control, in Figures 1c and d were published incorrectly. The experiment was repeated and the correct versions of these figures and figure legends are shown below.
(c) Both TNF and EGF induce IκBα degradation. Cells were treated with 0.1 nm TNF or 100 ng/ml EGF for the indicated times and then cytoplasmic extracts were prepared and western blot analysis was performed with anti-IκBα antibody. (d) TNF induces IκBαphosphorylation at the Ser32/36 position, but EGF does not. Cells were preincubated with 100 μg/ml proteasome inhibitor ALLN for 1 h and then treated with either 0.1 nm TNF for 15 min or 100 ng/ml EGF for 60 min. Cytoplasmic extracts were prepared and then western blot analysis was performed with phospho-specific anti-IκBα antibody.
We’ve reached out to Aggarwal, the editors of Oncogene and the International Journal of Cancer, and MD Anderson. We’ll update this post with anything else we learn.
Hat tip: Jaime A. Teixeira da Silva and anonymous
Like Retraction Watch? Consider making a tax-deductible contribution to support our growth. You can also follow us on Twitter, like us on Facebook, add us to your RSS reader, and sign up on our homepage for an email every time there’s a new post. Click here to review our Comments Policy.
I’m confused.
“In each case, the same blot was stripped and reprobed with anti-b-actin antibody to show equal protein loading (lower panel in each figure)”
and
“Beta actin controls (for different cell lines) in panel (a), (b) and (e) were derived from the same experiment. Beta actin controls (for different curcumin doses) for panel (a), (b), (d) and (e) were also derived from the same experiment.”
These statements seem to contradict one another. Also, are they suggesting that since the beta-actin blot was on the same samples (and not the same blot), it is o.k. to use the same image as a loading control? Have I been doing loading control wrong all of this time?
It looks like there now are a lot of retractions (at least four by my count) – I assume RW will be covering them soon?
We’ve reported on this case since this post: http://retractionwatch.com/2016/02/22/journal-retracts-7-papers-by-md-anderson-researcher-long-under-investigation/ Let us know if you see other retractions.
Ah, sorry, missed that one. It didn’t show up when I searched for Aggarwal’s name in your search field.