A UPenn professor now has six papers with a correction, expression of concern, or retraction in two PLOS journals after one published an extensive correction to a 2018 paper.
The correction adds to two retractions and three expressions of concern for papers in PLOS Pathogens and PLOS ONE with Erle Robertson, a microbiology professor and vice chair of research for the department of otorhinolaryngology at the University of Pennsylvania, as a senior author. The actions on each paper happened after commenters on PubPeer pointed out issues.
The correction to “STAT6 degradation and ubiquitylated TRIML2 are essential for activation of human oncogenic herpesvirus” states:
The Fig 5B K6-STAT6-Ub (HA) panel is incorrect; instead of the correct K6 panel, an image of the WT results at a shorter exposure time was used inadvertently. Fig 5 has been updated below to show the correct K6 panel. The underlying image data for the blots presented in Fig 5 are provided in the S1 File below.
The Fig 7A iSLK-KSHV GAPDH panel is incorrect, and Fig 7 has been updated to present the correct iSLK-KSHV GAPDH panel. The underlying image data for the blots presented in Fig 7 and the underlying individual level data presented in Fig 7E are provided in the S1 and S2 Files below.
In addition to the errors above, the BC3 STAT6, BCBL1 STAT3, and BC3 GAPDH panels of Fig 1A have not been prepared in line with best practice guidelines for image preparation. These panels have been replaced in the updated Fig 1 below to represent the obtained results more accurately. The underlying image data for the blots presented in Fig 1 are provided in the S1 File below. The individual level data underlying the results presented in Fig 1D, 1E, 1F and 1G are provided in the S3–S6 Files below respectively.
Furthermore, the authors would like to clarify that the similarities between the Fig 1A BCBL1 STAT6 panel and the Fig 4B BCBL1 STAT6 panel, as well as the similarities between the Fig 1A BCBL1 GAPDH panel, Fig 4B BCBL1 GAPDH panel, and the Fig 7B BCBL1 GAPDH panel are due to these panels being used to represent results obtained from the same experiments. The underlying image data underlying the results presented in Figs 1, 4 and 3 are provided in the S1 File below.
The paper has been cited 10 times, according to Clarivate’s Web of Science.
A PubPeer commenter had noted in January 2020 that aspects in figure 5B were “much more similar than you would expect.” Another wrote in May 2020 that Robertson appeared to be on the editorial board of PLOS Pathogens, but he is not currently listed on the editorial board page.
PLOS spokesperson David Knutson confirmed:
Dr. Robertson’s role as a section editor ended in April 2021. We don’t generally comment on editor departures for privacy reasons.
Robertson and Qiliang Cai, the corresponding author of the paper from Fudan University in Shanghai, did not respond to our request for comment.
We asked Knutson about the decision to correct the paper rather than issue an expression of concern or retraction. He said:
PLOS abides by the relevant COPE guidance and the PLOS policy on Corrections, Expressions of Concern, and Retractions when deciding how to resolve post-publication cases. The reasons for the editorial decisions taken for this series of articles are explained in the published notices. We stand by all of the decisions.
For ppathogens.1007416, the correction may seem extensive, but the central concern with the article regarded an error in Figure 5B and one in Figure 7A. The rest of the notice was for clarification purposes.
According to our database, five of Robertson’s other papers in PLOS Pathogens and PLOS ONE, three with Qiliang as a coauthor, have been flagged with expressions of concern or retracted since last April:
- “KSHV-Mediated Regulation of Par3 and SNAIL Contributes to B-Cell Proliferation,” from PLOS Pathogens in 2016, expression of concern dated April 11, 2022 (cited 20 times)
- “Ubiquitin/SUMO Modification Regulates VHL Protein Stability and Nucleocytoplasmic Localization,” from PLOS ONE in 2010, retraction dated March 4, 2022 (cited 13 times)
- “Nuclear Localization and Cleavage of STAT6 Is Induced by Kaposi’s Sarcoma-Associated Herpesvirus for Viral Latency,” from PLOS Pathogens in 2017, expression of concern dated December 15, 2021 (cited 15 times)
- “Early Events Associated with Infection of Epstein-Barr Virus Infection of Primary B-Cells,” from PLOS ONE in 2009, expression of concern dated August 19, 2021 (cited 68 times)
- “Epstein-Barr Virus Nuclear Antigen 3C Stabilizes Gemin3 to Block p53-mediated Apoptosis,” from PLOS Pathogens in 2011, retraction dated April 30, 2021 (cited 52 times)
Each paper has either Robertson or Qiliang as the corresponding author. The notices all similarly list concerns raised with multiple figures and the authors’ responses, and the expressions of concern include additional data the authors submitted.
The PLOS ONE retraction notice cites “unresolved concerns about image data reporting and the reliability of results” in one of the figures, and notes that Robertson and Qiliang did not agree with the retraction. The PLOS Pathogens retraction notice similarly attributes the decision to “concerns affecting multiple figure panels that question the integrity of these data.” It states that Robertson and Qiliang “agreed with the retraction, but stand by the article’s findings.”
We asked Knutson about what happened to spark the action on these papers. He said:
Readers shared some of the concerns with these articles directly with the journal. During our initial assessment, PLOS also became aware of concerns shared on PubPeer and on Social Media platforms.
PubPeer comments on the articles, some by data sleuth Elisabeth Bik, date back to 2020.
On the 2016 PLOS Pathogens paper, Bik noted concerns beyond the usual image duplication:
Figure 8B.
Could the authors please comment on the size of some of these tumors [in mice]? They appear to be very large.
The Methods state that “The office of Laboratory Animal Welfare (OLAW) guidelines provided by National Institute of Health (NIH) USA, are strictly followed by the University of Pennsylvania Institutional Animal Care and Use Committee (IACUC). Our protocol number for this study was 804617 and we have adhered with all OLAW guidelines.”
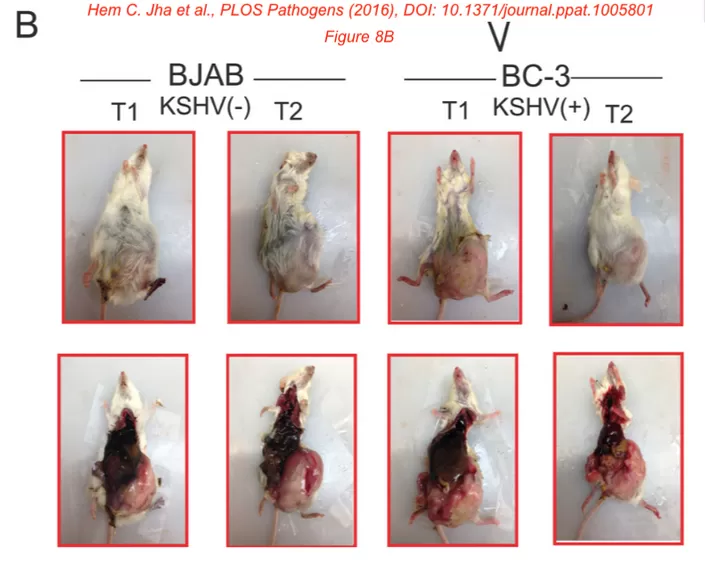
The expression of concern on the paper contains the response from the corresponding author (Robertson) to this and other concerns, and the journal’s ruling:
The corresponding author disagrees with the concerns raised regarding the tumor sizes reported in Fig 8B. They state that the animals used in this study were monitored daily and indicated that they would immediately proceed with euthanising if the weight of the mouse with tumor growth and ascites exceeded 20% of the initial weight, or if any of the animals were in distress at any point. However, the authors did not provide individual level data on tumor measurements requested by the journal, but instead the corresponding author stated that sometimes it is possible that a 40 to 50% increase in tumor size may occur within a 24-hour interval. In the absence of individual level data reporting the tumor measurements the journal remains concerned that the tumors reported Fig 8B exceed sizes commonly accepted for mouse tumor studies. The authors have not provided underlying data files to support the results presented in Fig 8.
The PLOS Pathogens Editors issue this expression of concern to notify readers of the above issues.
Like Retraction Watch? You can make a tax-deductible contribution to support our work, follow us on Twitter, like us on Facebook, add us to your RSS reader, or subscribe to our daily digest. If you find a retraction that’s not in our database, you can let us know here. For comments or feedback, email us at [email protected].

This is well needed, welcome input from PubPeer and other social media platforms. So many journals, open source with requests for rushed reviews has let errors and weaker science get published. I’m very glad to see the retractions.
Partially due to the current academic system, professors have no time to look into data from close colleagues (e.g. postdocs) as they have to spend so much time on grant applications to survive, particularly those in medical schools. Is there a way to improve the system? How to create a helpful lab environment might be the best approach we can think of.
Some manage without publishing problematic data.
There is likely a myriad of reasons for problematic data, but the important part is to correct, or retract, the problematic data.
This author has papers with concerns going back 20 years. https://pubpeer.com/search?q=authors%3A%22Erle+s.+Robertson%22
Here’s a link to a problematic image from a 2004 paper, which looks like a valid concern to me: https://pubpeer.com/publications/11B2A7805D5C1BF6872C8721870ED9#2
How many years back should letters of concern or requests for retraction be made? How about 50 or 100 years? Does truth matter?
That’s at least an order of magnitude short.
https://en.wikipedia.org/wiki/Donation_of_Constantine