
Ever wanted to hone your skills as a scientific sleuth? Now’s your chance.
Thanks to the American Society for Biochemistry and Molecular Biology (ASBMB), which is committed to educating authors on best practices in publishing, figure preparation, and reproducibility, we’re presenting the thirteenth in a series, Forensics Friday.
Take a look at the image below, and then take our poll. (We recommend using the Chrome browser.) After that, click on the link below to find out the right answer.
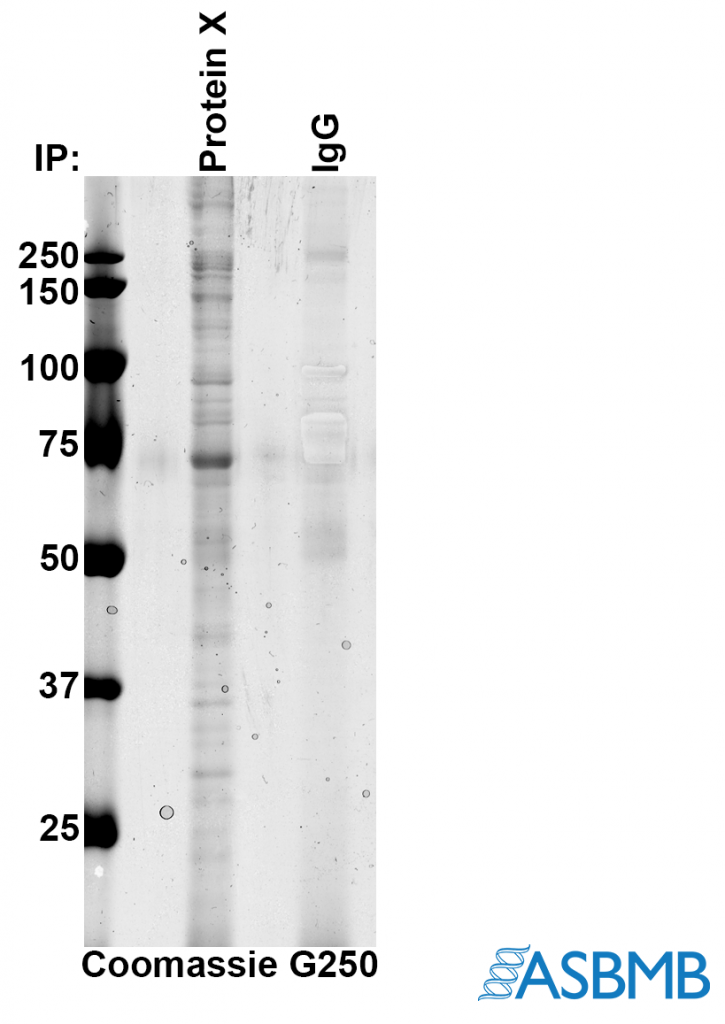
Think you chose the right answer? Click here to find out.
And check out the last Forensics Friday.
Like Retraction Watch? You can make a tax-deductible contribution to support our work, follow us on Twitter, like us on Facebook, add us to your RSS reader, sign up for an email every time there’s a new post (look for the “follow” button at the lower right part of your screen), or subscribe to our daily digest. If you find a retraction that’s not in our database, you can let us know here. For comments or feedback, email us at [email protected].
I use Chrome browser and still cannot see the quiz choices for Forensic Friday; it does not work properly.
This is a great series RW. And useful! But reviewers see more than blots. Science is lots more than that. How about some questionable FACS data, single channel recordings, Fura traces, and much, much more .. . you name it?
Glad you like Forensics Fridays!
We’d be happy to include other kinds of images, but these are a partnership with ASBMB, as the copy notes, and these are the kinds of images they can provide. We’re very grateful for their help, and we don’t have the resources to develop these on our own. If you have suggestions of who might be able to help us — including yourself, given your experience! — we’re all ears.
I have divers examples from multiple powerpoints that were publicly delivered in many venues (meaning they were in public domain) that I would happily volunteer. We could talk offline about any issues, etc. Some I purposely falsified de novo for teaching purposes.
Surely others, maybe current RCR course instructors, would be able to generate more modern examples?
Elisabeth Bik has an endless catalog of examples of *real-life* faked images, in varying levels of transparency, but you are probably looking for *simulations* of fakery to avoid the complications of copyright and reputation-protection lawyers.
Hmm, seems to me RW has got it wrong again. The problem you identify isnt the main one. (yes what they did is wrong but there is a more fundamental technical problem)
The biggest issue for me is how come the IgG isnt more obvious. The band at ~70 kD is much more intense than the IgG,and since its coomassie, it means its much more abundant. For that to be real, there would have to be loads of the 70kD proteins bound per molecule of protein X.
More likely explanation – the band at 70 kD is BSA.
Alan, I thought IgG was the control lane — as in, a control IP with just purified IgG instead of anti-protein X on the beads/resin.
In that case, the authors may have been selectively enhancing to show just how little came down in the control at those sizes… but it’s not clear why they wouldn’t just burn in the whole lane or (better) cut it out of the gel and silver stain it.
Also, those don’t look so much like selective enhancements as some sort of bubble… and they are absent in the pdf that RW presents in the “answer”, so it’s difficult to compare with the png above. (Who is manipulating images here exactly?) I took screenshots, which can be seen here: http://sennoma.fastmail.fm/Capture.PNG.
Finally, I’d need to know how they got the co-IPs off the beads, since there are no obvious H+L chain bands in the control.