
Ever wanted to hone your skills as a scientific sleuth? Now’s your chance.
Thanks to the American Society for Biochemistry and Molecular Biology (ASBMB), which is committed to educating authors on best practices in publishing, figure preparation, and reproducibility, we’re presenting the eleventh in a series, Forensics Friday.
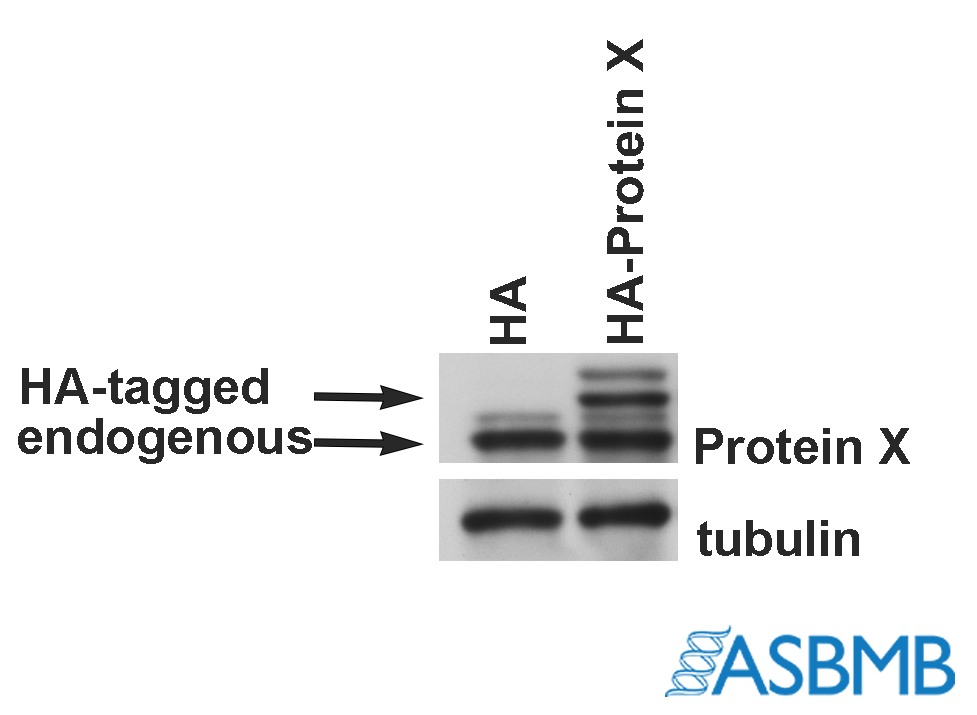
Take a look at the image below, and then take our poll. (We recommend using the Chrome browser.) After that, click on the link below to find out the right answer.
Think you chose the right answer? Click here to find out.
And check out the last Forensics Friday.
Like Retraction Watch? You can make a tax-deductible contribution to support our work, follow us on Twitter, like us on Facebook, add us to your RSS reader, sign up for an email every time there’s a new post (look for the “follow” button at the lower right part of your screen), or subscribe to our daily digest. If you find a retraction that’s not in our database, you can let us know here. For comments or feedback, email us at [email protected].
This is potentially an interesting exercise, but without a lot more context it’s hard to say what additional controls are necessary. It’s conceivable that this data demonstrates sufficient antibody specificity for a narrow set of downstream experiments – maybe everything is done by overexpressing Protein X-HA, using the HA tag to immunoprecipitate, and using this new antibody for blots.
Of course, more controls are usually a good idea. If downstream experiments are used to analyze endogenously expressed protein in some cell or tissue of interest, it becomes critical to demonstrate specificity by knockdown/knockout. Ideally in the exact sample being analyzed, but that can be infeasible or impossible.
(I’ve always used null mutants to validate the antibodies I’ve made, but that’s with the luxury of model organism genetics and stock centers. We can’t expect every mouse researcher to make a knockout just to validate their antibody, and there’s no way to do the knockdown/knockout in human primary tissue samples)
Full blots should also be provided as a matter of course, but that’s something I’d probably leave to the journal editor to enforce their submission guidelines. But again, context matters. If later experiments rely on the antibody to detect cleavage products, dimers, or anything else of a different molecular weight, showing this blot uncropped is absolutely critical.