In March 2017, a group of researchers in Vancouver, along with a colleague in Philadelphia, published a paper in the Proceedings of the National Academy of Sciences (PNAS) concluding that a particular antibiotic might be useful for treating conditions in people with rare mutations.
Then, this past July, while continuing the work, they had an unexpected result. That made them suspect that the antibiotic they thought they had ordered, gentamicin, wasn’t what they thought it was. With the help of a different company that sells the antibiotic, they confirmed they were studying a different compound — and retracted the paper.
Here’s the notice for “Gentamicin B1 is a minor gentamicin component with major nonsense mutation suppression activity:”
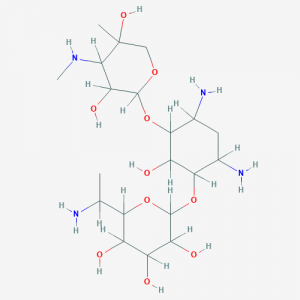
We have now determined that the gentamicin B1 compound we acquired commercially and used in our study was not gentamicin B1 but the closely related aminoglycoside G418. At the time we carried out the work, the only commercial source for gentamicin B1 was MicroCombiChem. The company provided two batches of compound, purified from gentamicin sulfate complex c, with certificates of analysis verifying the compound to be >97% pure gentamicin B1, together with HPLC-MS and NMR analysis showing the data conformed to gentamicin B1. Moreover, we asked a third party to carry out independent NMR analysis and they determined that the MicroCombiChem compound had a nitrogen in the vicinity of a methyl group, which is the case for gentamicin B1 but not for G418.
In July 2018, while carrying out structure–activity relationship studies, we observed that a newly synthesized aminoglycoside derivative containing ring 1 of gentamicin B1 unexpectedly lacked nonsense suppression activity. This finding made us suspect the nature of the compound purchased from MicroCombiChem. At this time, gentamicin B1 became available from a second commercial source—Toronto Research Chemicals. Chemistry collaborators David Powell, David Williams, and Raymond Andersen graciously agreed to carry out NMR analysis of G418 disulfate salt from Sigma, gentamicin B1 free base from MicroCombiChem and gentamicin B1 diacetate salt from Toronto Research Chemicals. They determined that gentamicin B1 provided by MicroCombiChem was G418 whereas gentamicin B1 provided by Toronto Research Chemicals was composed principally (>95%) of gentamicin B1.
We note that gentamicin B1 and G418 are closely related compounds, having the same atomic composition and molecular mass but differ in the location of amine and hydroxyl functionalities in ring 1. Ascertaining the structure of gentamicin B1 and identifying the NMR signals that distinguish it from G418 was not a trivial task because there are no published NMR data for gentamicin B1, and it was observed that the resonances of methylene protons vicinal to amino nitrogens in G418 and gentamicin B1 shifted considerably depending on pH and solvent. This could have led to the misidentification of G418 as gentamicin B1. Details of the NMR and biological analyses have been deposited in bioRxiv.
All biological data presented in the PNAS paper (Figs. 1, 3, and 4, and Figs. S1, S3, S4, and S5) are accurate except that the results attributed to gentamicin B1 should now be considered to pertain to G418. We wish to apologize to the scientific community for any adverse consequences that may have resulted from following our work. Accordingly, we hereby retract the article.
The paper has been cited 17 times, according to Clarivate Analytics’ Web of Science. MicroCombiChem did not respond to requests for comment.
We asked Anita Bandrowski, who recently wrote about a catalog typo that led to a correction, for her take. She dug into the case, and sent us some comments:
Neither Gentamicin B1 nor G418 are currently listed for sale from MicroCombiChem. They have different PubChem CIDs: 3034288 for Gentamicin B1 and PubChem CID 123865 for G418.
The two molecules have the same molecular formula and molecular weight, so they are very similar, but clearly have different biological activity. This is not uncommon with chemical, even enantiomers where one can have a beneficial drug like activity and the other can be inert.
Currently we do not know how many manufacturers have a problem distinguishing between these two compounds and therefore we are unable to understand how this affects the scientific literature.
We do know that “gentamicin B1” is discussed in 114 papers according to Google Scholar, however there are ten different approved names for this compound, according to PubChem, therefore searching in the literature for all of those names as well as the variants and misspellings would be tricky. None of the papers refer directly to the PubChem CID for this compound as something that the authors used, making the search task even more difficult.
It would be quite interesting to understand if other manufacturers of this reagent are similarly affected, and it would be interesting to understand how many papers may need to be corrected or retracted because of this issue and because there are no clear and easy ways to do this.
So would have Bandrowski’s initiative designed to help researchers identify their reagents correctly have prevented this mishap?
From our perspective at the [Research Resource Identifier] RRID project, this case demonstrates the dire need for all reagents, or at least tricky reagents like antibodies, to be tagged with RRID identifiers because that provides information on the things that authors actually used in their study in a very easy to search format. The easy to search part is important because a machine could in theory run over this complete literature and flag papers where a potential problem was detected, but the myriad of ways that a reagent can be described makes this sort of detection very difficult.
Like Retraction Watch? You can make a tax-deductible contribution to support our growth, follow us on Twitter, like us on Facebook, add us to your RSS reader, sign up for an email every time there’s a new post (look for the “follow” button at the lower right part of your screen), or subscribe to our daily digest. If you find a retraction that’s not in our database, you can let us know here. For comments or feedback, email us at [email protected].
By the sounds of the retraction notice, an RRID wouldn’t have done anything to help these researchers.
I use drugs in my research all the time. I just have to suck it up and hope that the tiny bits of powder they send me are the drugs that are on the label, knowing full well that a portion of the time that they might not be. To their credit these authors sent it off for NMR verification which came back OK.
SciCrunch does not provide RRIDs for drugs and reagents so the researchers wouldn’t have been helped by an RRID. They still would have trusted the company to send them the correctly labelled product and still would have trusted their initial NMR analysis.
In fact, there are good reasons why RRIDs are not necessary for drugs and chemicals. We already have at least several good standards that define these chemicals in a unique way. Many papers give the long IUPAC chemical name – that’s 1 standard. But if they wanted to save a bit of space, they could just give a pubchem ID, an IUPHAR ligand ID, a CAS number, or the chemical structure in SMILES.
Seriously, an extra ID number isn’t going to help if the label on the bottle is wrong or if your compound decays into a closely related isomer over time and forgot to relabel itself.
if your compound decays into a closely related isomer over time and forgot to relabel itself.
http://retractionwatch.com/2017/12/04/caught-notice-find-paper-relied-expired-material/
I have to say, I really really like the concept of self-relabeling containers (I don’t think it would be fair to put all the burden on the contents, be they compounds or not). Just as one other example, such a technology (done really well) would finally allow one to fairly judge a book by its cover!
Here’s a feature article I wrote for Nature last year that has some similar stories of chemical mix-ups, including some problems with gentamicin. https://www.nature.com/articles/548485a
These authors did do one of the main and oft-neglected steps for quality control, NMR. Tricky stuff this.
https://www.nature.com/articles/548485a