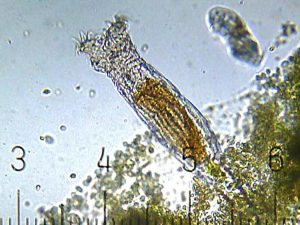
A group of tiny, all-female animals called bdelloid rotifers has long fascinated scientists. Among other questions, of course, is: Why haven’t they gone extinct, if they can’t mix up their genes? In 2016, a group of authors published a paper in Current Biology claiming to show that rotifers could swap DNA the way bacteria do. But a paper published earlier this month in the same journal found “clear evidence that the data and findings of [that study] are unreliable.” It’s unusual for a journal to publish a full paper clearly refuting another — and Current Biology left the original paper as is, without even a link to the new one. The authors of the new paper — Chris Wilson, Reuben Nowell, and Tim Barraclough, all of Imperial College London — explain how all of this came to pass, and why the authors of the original paper deserve praise.
RW: You and others refer to asexual rotifers as an “evolutionary scandal.” Can you explain?
Chris Wilson, Reuben Nowell, and Tim Barraclough (CW, RN, TB): Sex is a fundamental puzzle for biologists. It’s a very inefficient way to make offspring, but nearly all plants and animals do it. Groups that abandon sex and start cloning themselves quickly go extinct, and we don’t fully understand why. There is something vital about shuffling up DNA.
We and our colleagues study tiny aquatic animals called bdelloid rotifers. They have diversified into hundreds of species over millions of years, but no males are known and the females reproduce via unfertilised clonal eggs. Without sex and genetic shuffling, these animals ought to have gone extinct, but instead they are doing fine. Thirty years ago, a prominent theoretician called the bdelloids “an evolutionary scandal,” and they are still causing trouble today.
In 2016, a paper in Current Biology seemed to show that these rotifers make up for their lack of sex by swapping DNA with each other horizontally, like bacteria do. No other animals are known to do this, but there was speculation that bdelloids might, because we know they have acquired many genes horizontally from bacteria, plants and fungi over millions of years. It’s important to note that horizontal gene transfer from bacteria, plants and fungi into bdelloid rotifers is a clear and well–established finding. It’s not an artefact of contamination (cf. #tardigate), and it’s not what we’re talking about in the new paper. We address a specific 2016 paper, which claimed to show that bdelloids are also swapping DNA horizontally with each other.
RW: You found that the data and findings of the previous study were unreliable. What do you think should happen to the paper?
CW, RN, TB: Our new paper in Current Biology provides “clear evidence that the data and findings of Debortoli et al. (2016) are unreliable.” The results are determined to be artefacts caused by “accidental cross-contamination between tubes during preparation of samples”. They are found to offer “no credible evidence for horizontal DNA exchange within or between bdelloid species.” This isn’t just an alternative interpretation, or a failure to replicate — we showed the DNA contamination directly in the original raw data from the 2016 paper.
As we understand it, decisions about the status of a paper rest firstly with its lead authors and then with the editors of the journal, who have final responsibility. We would guess they are considering this case, but it’s not for us to advise them what to do. Our role was to notice a problem and present the evidence for our hypothesis of contamination: first to the original authors, then to editors and reviewers at Current Biology and now to the wider scientific community.
We hope that readers of our paper will weigh the evidence and share their thoughts with others. We’d like the message to reach colleagues who might otherwise cite the previous paper or spend time and effort trying to build upon its findings. One helpful step would be for Current Biology to provide a visible link to our paper at the DOI for the 2016 publication. They have told us they will try to do this if technically possible.
RW: The last author of the 2016 paper told The Scientist that “It’s not clear from the data they show that this really affects our analysis.” Do you agree?
CW, RN, TB: No, and it is not entirely clear they still agree with that themselves. They have since posted a preprint on bioRxiv, in which they apply a useful automated version of our statistical technique to their own data. Their analysis reveals even more extensive cross-contamination than our manual tests had found. They write that “we cannot rule out at this stage that some of the signatures of interspecific recombination reported in our 2016 article actually resulted from contamination”. They note that DNA contamination from various sources is a common and well-known issue in many molecular laboratories. We agree with this, but the specific problem with their work is that cross-contaminated DNA samples were exponentially amplified and used to support biologically extraordinary claims about horizontal DNA transfers. Since both groups now independently report pervasive cross-contamination in the 2016 samples, it seems clearer than ever that the data and conclusions about horizontal transfer of DNA are not reliable.
At the end of their preprint, the authors write that it remains an open question whether or not bdelloid rotifers exchange genes in unusual ways. We agree, and we look forward to future data that will throw more light on these troublesome creatures. As our colleagues remarked in the same interview, this exchange is a good thing for science in the long term. It is already generating techniques and insights that can be applied more broadly.
RW: You write in the acknowledgements that “We appreciate their transparent and scholarly conduct and their thoughtful and collegial correspondence after we first communicated our concerns in April 2016.” Can you say more about what happened? Was it all smooth sailing with the editors, too?
CW, RN, TB: We first noticed a puzzle soon after the paper appeared. All the reported donors and all the recipients of “horizontal DNA transfer” occurred in the same small sample of animals that the authors sequenced. This suggested cross-contamination rather than a biological signal, but was not conclusive. A stronger clue came in April 2016, when the genetic sequences were released at GenBank. DNA molecules that had supposedly paired up and swapped sequences were only about 70% identical to each other. According to measurements from other organisms (even bacteria), homologous recombination does not occur between such vastly different molecules, as they can’t pair up properly. Again, this pointed to a contamination artefact, not true exchange.
We shared these two clues with the authors right away. We suggested the hypothesis of DNA cross-contamination, and asked if it might be visible in their raw sequencing chromatograms. They thanked us very cordially for our input. They took our concerns seriously: they agreed it was a real and worrying possibility that small amounts of contaminating DNA might have been amplified and “swamped” the signal from the bona fide sequence. They wrote that the easiest way to test the hypothesis would be to examine the chromatograms for small double peaks (even tiny ones) that could indicate contamination. In November 2016, they kindly offered to share chromatograms with us, and sent the key files within 24 hours. This was very helpful, as raw DNA sequencing data of this sort are rarely deposited alongside edited genetic sequences.
The chromatograms were noisier than expected, especially for the supposed recipients of horizontal transfer. After looking closely, we found many cases where small secondary peaks seemed to match the sequence of the apparent DNA “donor”. We highlighted some of those peaks to our colleagues, and let them know we thought the data confirmed cross-contamination. After careful consideration, they disagreed with this interpretation, and took the time to explain their reasoning in detail. They suggested we might be seeing patterns we expected to see in the considerable noise, and that the extra peaks might not be meaningful, because they were quite small.
We carefully considered this in turn, as it is very common to see patterns that confirm a previously held belief. Over several months, we developed a simple statistical method, described in our paper, to test for a consistent pattern of extra peaks. We addressed the question about the size of peaks experimentally: we deliberately put two different rotifers in the same tube, extracted DNA and amplified it using the methods of the 2016 paper. As predicted, the extra peaks were small, but the statistical method detected the deliberate contamination with high confidence. When we applied our validated method to the 2016 data, we found clear evidence of cross-contamination in samples showing “horizontal genetic exchanges”.
At that point, two of us were due to speak about asexual rotifers at an international conference. It would have been difficult not to mention the prominent 2016 paper, or to do so without sharing our concerns and new results. However, it did not seem right to drop this in a passing slide or during conference chatter, without providing the community with evidence to evaluate our contamination hypothesis. We decided to post a preprint at bioRxiv.org. We let the original authors know, and shared an advance copy. From that point, the matter is in the public record, including a further helpful comment from our colleagues on the preprint that led to an extension of the manuscript.
When we decided to approach Current Biology, the editors responded positively and quickly, in full knowledge that our work directly concerned the reliability of a paper they had published. They initially thought we might write a short correspondence piece instead of a full paper, but after considering the data and detail we had to present, we agreed on the latter. We are grateful to the editors for being receptive, and to three reviewers for constructive and supportive comments that improved the paper. We also thank the original authors again for their collegial correspondence and sharing of data.
RW: What would you tell researchers interested in correcting the scientific record they should expect, based on your experience?
CW, RN, TB: It’s important to note that our experience relates to a case of inadvertent error, and our colleagues engaged with us in an open and constructive way, even when resisting our conclusions. Other situations might unfold differently.
Patience and perseverance are crucial. Like all scientific projects, investigating a possible error in published work takes time and care. Any authors will understandably be strongly invested in defending their work, and it is important to be considerate in giving them the opportunity to do so. If they are willing to engage, they may devote considerable time and attention to locating flaws in the critique and gaps in the evidence. Quantitative rigor is rightly insisted upon, perhaps even more so than in other projects. If all peer reviewers were so strongly motivated to scrutinise details and probe arguments, perhaps the scientific record would contain fewer errors and replication failures.
This case is also a little unusual. Contamination may be the most common source of honest error in the retracted literature, but it is hard to show that someone else’s findings are unreliable for that reason. One might raise it (cautiously) as an alternative interpretation, but it’s rarely possible to demonstrate contamination in the same raw data as the original study, as we can with DNA. Even in this case, the signal could easily have been lost. If the authors had used a slightly different method (cloning the DNA before sequencing), there would have been no extra peaks and no way to reconstruct the error. Other clues about contamination would remain, but clear evidence to resolve the point would be lacking. Sometimes, no matter how confident you are that an error has been made, or how much time you spend investigating, the critical data to test the hypothesis directly might not exist, especially if the original reagents or samples have been consumed. The best you can do is to show that the work does not replicate. This can be frustrating for scientists, as we are accustomed to getting answers out of natural systems if only we are clever and persistent enough. It’s important to recognise that engaging with colleagues brings a different set of considerations.
Like Retraction Watch? You can make a tax-deductible contribution to support our growth, follow us on Twitter, like us on Facebook, add us to your RSS reader, sign up for an email every time there’s a new post (look for the “follow” button at the lower right part of your screen), or subscribe to our daily digest. If you find a retraction that’s not in our database, you can let us know here. For comments or feedback, email us at [email protected].