A paper on the genetics underlying a common neurological disorder has issued a correction that influences the results of the paper.
“Genetic Diagnosis of Charcot-Marie-Tooth Disease in a Population by Next-Generation Sequencing” was published in BioMed Research International, and looked at 81 families with the disease. The researchers identified mutations that might be connected to the disease. The problem, says the correction note, is that the authors classified a couple variants in one patient’s genes as “likely pathogenic,” when their true nature was less clear.
The correction explains the new numbers:
In Table 4 and Supplemental Table 1 of the published paper entitled “Genetic Diagnosis of Charcot-Marie-Tooth Disease in a Population by Next-Generation Sequencing” [1], we classified two variants as likely pathogenic. These two variants (NM_001126131.1:c.[1491G>C(;)2243G>C]), identified in the POLG gene, were carried by one patient (ID 62). As it was not possible to examine whether the two variants were in cis or trans, that is, situated on the same or different alleles, these two variants should have been reported as uncertain pathogenic variants.
This change also influences the Results, where the prevalence of identified variants is corrected to 8 likely and 17 uncertain pathogenic variants.
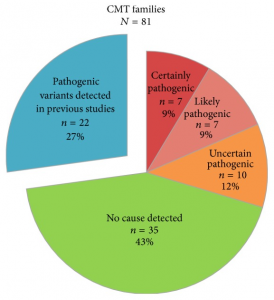
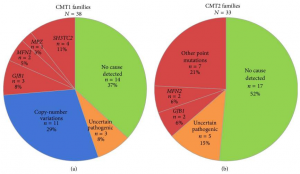
In Figure 1, the number of families with likely pathogenic variants is reduced to n = 7 (9%) and the number for families with uncertain pathogenic variants is increased to n = 10 (12%). And in Figure 2(b), the number of CMT2 families in the category “other point mutations” is reduced to n = 7 (21%) and the number of families with uncertain pathogenic variants is increased to n = 5 (15%). See corrected Figures 1 and 2(b).
Here’s the corrected Figure 1:
Identified variants in 81 Norwegian CMT families from the general population. Our previous studies identified copy-number variations in 12 CMT families and pathogenic point mutations in 10 CMT families [2, 14, 19]. The remaining 59 CMT families were investigated by next-generation sequencing.
And Figure 2:
Frequencies of certain and likely pathogenic variants in CMT1 and CMT2 families from the Norwegian general population.
We asked Hindawi Publishing Corporation, the publisher of the journal, if they had considered retracting the article. Publishing Editor Sarah Ahmed told us:
Our decision to issue a correction rather than a retraction for this article were based on COPE’s guidelines as well as the decision of the academic Editor in charge of this article.
The Committee on Publication Ethics guidelines suggest issuing a correction if:
…a small portion of an otherwise reliable publication proves to be misleading (especially because of honest error)
We’ve reached out to last author Michael Bjørn Russell at the University of Oslo, and first author Helle Høyer, a student at Sykehuset Telemark at the time that the work was done. We will update this post with anything else we learn.
Like Retraction Watch? Consider making a tax-deductible contribution to support our growth. You can also follow us on Twitter, like us on Facebook, add us to your RSS reader, and sign up on our homepage for an email every time there’s a new post. Click here to review our Comments Policy.
I think Retraction Watch performs a valuable service to the scientific community (and beyond), so I apologize that my first comment is a mild reproach. This story is just not newsworthy. A small error has been made and has been rightfully and honestly corrected. Is reporting on something as trivial as this really how you guys want to spend your time? Beyond the insignificance of the report as a news item, it only serves to draw negative attention to a group who has honestly, even courageously, set the record straight. Reporting on this action, and further insinuating that the proper course of action would have been a retraction, suggests that you are more interested in getting mud to stick than shining a light. Please think twice, even three times, before reporting on honest corrections of a trivial nature.
A paper on the genetics underlying a common neurological disorder
I do not think this is true, except in comparative terms. Better, perhaps, to say that CMT syndrome (affecting “approximately 1 in 2,500 people” in the infallible words of the Whackyweedia) is less infrequent than many other neurological disorders.
This correction is highly commendable and to suggest retraction is absurd. I understand that you guys are not geneticists, but try to understand that these prediction algorithms are far from definitive anyway. They are at best guides to functional biologists like me about the possible impact of variants. We understand that they are to be taken with a grain of salt.
Coming from a reader not a geneticist; it appeared that it was not suggested that a retraction be made, the question was asked and the explanation given why a retraction was not necessary.