The journal Blood has issued a correction in a 2009 letter about the molecular underpinnings of chronic lymphocytic leukemia.
Despite the extent of the changes to a figure, “the error does not change the scientific meaning,” according to the erratum.
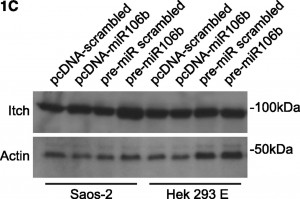
The article “p73, miR106b, miR34a, and Itch in chronic lymphocytic leukemia” was written in response to a 2009 Blood paper about the role of a microRNA in CLL. But its western blots were “assembled incorrectly,” leading to duplicated panels. Another set of panels was “shifted.”
So the authors repeated the experiments, and presented them in a correction. Here’s the correction notice in full, published earlier this month, including the figures in question:
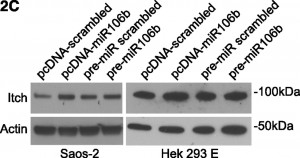
On page 6499 in the 18 June 2009 issue, there is an error in Figure 1C. The western blots are assembled incorrectly, featuring the same actin panel twice. Furthermore, the Itch panels are shifted. To further support the scientific conclusions, the authors repeated all experiments (Figure 2C, shown below), confirming the original results (corrected Figure 1C, shown below). Although the error does not change the scientific meaning, the authors apologize for the error.
Figure 1
Regulation of Itch in chronic lymphocytic leukemia. (A) Western blotting of expression levels of protein Itch. Primary chronic lymphocytic leukemia (CLL) cells were incubated in presence of 10 nM LBH589 for the indicated times. Levels of actin were used as a loading control. Results from a representative experiment using CLL cells from 1 of the 12 analyzed patients are shown. There is a decrease in the 100 kDa band but with concomitant increase in 64 kDa cleaved product. See also Rossi et al.3 (B) Luciferase reporter assay of miR106b on 3′UTR-Itch in Saos-2 and HEK293 cells. The results were expressed as mean + SD from 3 independent experiments analyzed in triplicate. (C) Western blotting of endogenous levels of protein Itch in Saos-2 and HEK293 cells upon transfection. Cells were transfected either with pcDNA plasmids harboring the mature miR106b or a scrambled sequence, or with pre-miRs for miR106b or a scrambled sequence. Levels of actin were used as a loading control. MiR106b levels were assayed by qRT-PCR, to verify the transfection. (D) Expression levels of TAp73, DNp73, miR34a, and miR106b in primary CLL cells exposed for the indicated times to 10 nM LBH589 as analyzed by qRT-PCR. Expression levels relative to the untreated sample at the corresponding time points are shown. Expression of TAp73 and DNp73 was normalized to actin; expression of miR34a and miR106b was normalized to RNU6b. Statistical differences were determined by one-way analysis of variance (ANOVA) followed by Dunnett multiple comparison test, and the results were expressed as mean ± SEM from 12 independent CLL samples analyzed in triplicate.
Figure 2C
A commenter on PubPeer first noted the problems with the figure in November 2013; a few others posted queries about why the issue took so long to resolve.
We contacted a couple of the authors involved. Corresponding author Martin Dyer at the University of Leichester in the UK sent us this response, back in June:
I have now partially retired and will not be able to reply to this email until July 12th when I return to the office.
I am sorry not to be able to offer more insight at this time
We followed up after July 12. We will update this post if replies.
We reached out to the journal for further comment, and communications specialist Amanda Szabo — from the publisher, the American Society of Hematology — got back to us with this response:
We hope that the explanation provided on the Blood website is self-explanatory and answers any questions.
Like Retraction Watch? Consider supporting our growth. You can also follow us on Twitter, like us on Facebook, add us to your RSS reader, and sign up on our homepage for an email every time there’s a new post. Click here to review our Comments Policy.

The senior author is editor-in-chief of Cell Death and Differentiation
http://www.nature.com/cdd/about.html#Editors
Pubpeer comments G Melino.
https://pubpeer.com/search?q=g+melino&sessionid=92ED1BC1695FCFF2732C&commit=Search+Publications
They have used a time machine. You cannot pin them down.
“To further support the scientific conclusions, the authors repeated all experiments [2013 or later] (Figure 2C, shown below), confirming the original results [2009] (corrected Figure 1C, shown below).”
Cell Cycle. 2013 Nov 15;12(22):3564-73. doi: 10.4161/cc.26771. Epub 2013 Oct 10.
GLS2 is transcriptionally regulated by p73 and contributes to neuronal differentiation.
Velletri T1, Romeo F, Tucci P, Peschiaroli A, Annicchiarico-Petruzzelli M, Niklison-Chirou MV, Amelio I, Knight RA, Mak TW, Melino G, Agostini M.
Author information
1Medical Research Council; Toxicology Unit; Leicester University; Leicester, UK; Institute of Health Sciences; Shanghai Institutes for Biological Sciences; Chinese Academy of Sciences & Shanghai Jiao Tong University School of Medicine; Shanghai, China.
2015 correction figure 1E.
http://www.tandfonline.com/doi/full/10.1080/15384101.2015.1035968
Did Martin Dyer ever reply?
Another Martin Dyer paper.
Cell. 1999 Jan 8;96(1):35-45.
Bcl10 is involved in t(1;14)(p22;q32) of MALT B cell lymphoma and mutated in multiple tumor types.
Willis TG1, Jadayel DM, Du MQ, Peng H, Perry AR, Abdul-Rauf M, Price H, Karran L, Majekodunmi O, Wlodarska I, Pan L, Crook T, Hamoudi R, Isaacson PG, Dyer MJ.
Author information
1Academic Department of Haematology and Cytogenetics, Institute of Cancer Research, Sutton, Surrey, United Kingdom.
Figures 4f and 4g.
http://i.imgur.com/B7E6XwA.jpg
https://pubpeer.com/publications/9989495